Inference of “ancestral recombination graphs” on a genome-wide scale.
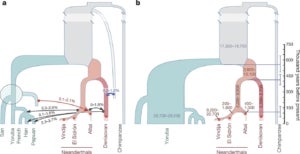
We have a longstanding interest in reconstructing the demographic history of complex, structured populations from DNA sequence data. Several years ago, Matt Rasmussen, a postdoc in the group, developed the first “ancestral recombination graph” (ARG) statistical inference method that is efficient enough to apply to complete mammalian genomes. We used this method, called ARGweaver, to detect gene flow from modern humans into the Altai Neandertal genome sequence and provided evidence to suggest an earlier migration of modern humans out-of-Africa than indicated by most current estimates. Later, Ph.D. student Melissa Hubisz extended ARGweaver to consider a full demographic model, including population sizes, divergence times, and migration events. Melissa then used this method to show that around 3% of Neandertal DNA—and possibly as much as 6%—came from modern humans who mated with Neandertals more than 200,000 years ago. She also predicted that 1% of the Denisovan genome introgressed from an unsequenced but highly diverged, archaic hominin ancestor. Intriguingly, about 15% of these “super-archaic” regions—comprising at least 4Mb—appear to have, in turn, introgressed into modern humans and continue to exist in the genomes of people alive today.
Related links:
- Our paper describing the ARGweaver method and its application to complete human genome sequences
- Our paper on modern human introgression into Eastern Neanderthals
- Our paper describing an extension of the ARGweaver method called ARGweaver-D
- Book chapter by Melissa Hubisz on the usage of the ARGweaver program
- Blog post by Adam Siepel on the ARGweaver method
Combining AI and ARG inference to study natural selection.
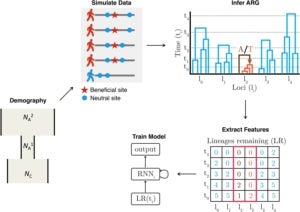
More recently, we have been finding ways of making use of the rich evolutionary information in reconstructed ARGs in other analyses, typically aided by methods from artificial intelligence. For example, in a study led by former postdoc Hussein Hejase, we used ARGweaver together with Hussein’s own machine-learning methods to study the genetic basis of speciation in a group of South American birds called Southern Capuchino Seedeaters. Working closely with collaborators Leo Campagna at the Cornell Laboratory of Ornithology and Ilan Gronau at the Herzliya Interdisciplinary Center in Israel, we were able to show that genealogical patterns at “islands of speciation” in these birds were more compatible with recent selective sweeps than with selection against gene flow. In particular, we found strong evidence of recent soft sweeps, many of which occurred near genes associated with plumage color, suggesting a prominent role for sexual selection in speciation. In a related project led by Hussein and Ph.D. student Ziyi Mo, we developed an improved machine-learning method called Selection Inference using the ARG (SIA), which refines predictions of selective sweeps and estimation of selection coefficients by taking advantage of a high-dimensional set of features extracted from inferred ARGs. Working again with Leo Campagna, Ziyi recently used SIA to characterize selective sweeps in another group of birds, Chestnut-bellied Monarchs from the Solomon Islands. Ziyi is currently extending SIA to take advantage of new “domain adaptation” methods borrowed from the image processing literature to be more robust to potential misspecification of the simulated data used for training. This strategy appears promising for many applications of machine learning that depend on simulated training data, in population genetics and other areas.
Related links:
- Our paper using the ARGweaver and machine-learning methods to study speciation in Southern Capuchino Seedeaters
- Our paper describing the SIA method
- Our paper on characterizing selective sweeps in Chestnut-bellied Monarchs from the Solomon Islands, in which SIA played a central role, with collaborators at Cornell Univerity
- Press release on our collaborative project with Cornell using SIA
Prediction of fitness consequences for mutations in humans and plants.
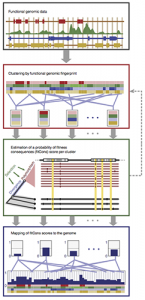
Another longstanding area of interest in the laboratory is the development of methods to predict the effects on evolutionary fitness of new mutations, in humans or other organisms. This work began with our development of a probabilistic model and inference method, called INSIGHT, that makes use of joint patterns of genetic polymorphism and divergence to shed light on the selective forces that have shaped genetic diversity. INSIGHT focuses in particular on disentangling the influences of positive and negative selection on collections of short, interspersed genomic elements, and we first used it to show that natural selection has profoundly influenced transcription factor binding sites across the human genome. Later, however, we realized that the same approach could be used to estimate the probability that a mutation occurring in any given collection of genomic sites will have fitness consequences. These “fitness consequences” (fitCons) scores turn out to be remarkably powerful for identifying cis-regulatory elements and are highly complementary to standard evolutionary conservation scores in revealing hidden functional elements. Building on this general idea, postdoc Yifei Huang and Ph.D. student Brad Gulko developed a series of methods that address variations on these problems—including LINSIGHT, which combines the INSIGHT model with a highly scalable generalized linear model, FitCons2, which simultaneously clusters and scores sites, and LASSIE, which makes use of the Poisson Random Field model to estimate allele-specific selection coefficients for potential mutations. Most recently, Yifei Huang, Noah Dukler, and Mehreen Mughal developed ExtRaINSIGHT, which takes advantage of patterns of rare variants in deep sequencing panels to estimate the prevalence of extremely strong purifying selection, or “ultraselection,” across the human genome. By applying ExtRaINSIGHT to more than 70,000 whole genome sequences from gnomAD, we found abundant evidence of ultraselection in evolutionarily ancient miRNAs and neuronal protein-coding genes, as well as at splice sites, but much less ultraselection in other noncoding RNAs and transcription factor binding sites, and only modest levels in ultraconserved elements. We have also been working with collaborators to apply these methods to newly generated data for agriculturally important plants, including rice, maize, and sorghum.
Related links:
- Our paper describing the INSIGHT method
- Our paper on INSIGHT and natural selection on human transcription factor binding sites
- Our paper describing the LINSIGHT method
- Our paper describing the LASSIE method
- Our paper introducing the fitCons approach and applying it in an analysis of three human cell types
- Our paper describing the FitCons2 model
- Our paper describing the ExtRaINSIGHT method
- Our paper using the fitCons approach and applying it to rice with collaborators at New York University
Analysis of nascent RNA sequencing data
Our research program in transcriptional regulation has focused on developing new methods for interpreting the rich nascent RNA sequencing (NRS) data generated using the powerful GRO-seq (Global Run-On and sequencing) protocol or its higher-resolution successor, PRO-seq (Precision nuclear run-on sequencing). Two recent projects have explored new uses of these data. First, postdoc Amit Blumberg developed a method for estimating relative RNA half-lives based on NRS data together with standard RNA-seq data. Amit’s method treats NRS read counts as a measure of transcription rate and RNA-seq as a measure of RNA concentration, and estimates the rate of RNA degradation required for steady-state equilibrium. He showed that this simple approach is remarkably effective, agreeing well with much more expensive and labor-intensive assays for RNA stability. Using this method, Amit showed that RNA splicing-related features are positively correlated with RNA stability, whereas features related to miRNA binding and DNA methylation are negatively correlated with RNA stability. He also identified several stability-associated histone modifications.
More recently, postdocs Yixin Zhao and Noah Dukler developed a new software tool called Deconvolution of Expression for Nascent RNA (DENR) to address the problem of pre-RNA isoform quantification. This is a critical technical problem that arises in almost all analyses of NRS data but, at best, it is typically addressed by heuristic filtering methods. DENR addresses the problem by modeling NRS read counts as a mixture of user-provided isoforms. A baseline mixture decomposition algorithm is enhanced by machine-learning predictions of active transcription start sites and an adjustment for the typical “shape profile” of read counts along a transcription unit. Yixin and Noah showed that DENR outperforms simple read-count-based methods for estimating gene and isoform abundances. They also showed that transcription of multiple pre-RNA isoforms per gene is widespread, with frequent differences between cell types. Indeed, they argued—based on an information-theoretic analysis—that a majority of human isoform diversity derives from primary transcription rather than from post-transcriptional processes.
Our current work in this area focuses in two main areas. First, we have a major collaborative project underway, with Charles Danko’s laboratory at Cornell, to collect PRO-seq data across the mammalian phylogeny, together with RNA-seq data, ATAC-seq data, and Hi-C/Micro-C data. The goal of this ambitious, multi-year project is to enable a comprehensive, multi-omic study of the evolution of gene expression in mammals. This project is an extension of a smaller study we published a few years ago, which focused on the evolution of primary transcription (using PRO-seq) in three primate species, and particularly on the evolution of enhancers and promoters in these species. Our second major ongoing project concerns the development of a “unified model” for analysis of NRS data, which includes both a kinetic model for the movement of RNA polymerases (RNAPs) across the DNA template and the generation of NRS read counts from underlying densities of RNAPs across populations of cells. This modeling framework has numerous applications, ranging from estimation of rates of initiation and promoter-proximal pause release, to tests for differences in such rates between species or cell types, to characterization of correlates (and potential causes) of variation in rates of elongation along genes. We have multiple active projects in this area and an NIH grant under review.
Related links:
- Our paper on estimating RNA half-lives using PRO-seq and RNA-seq
- Our paper describing the DENR method
- Our paper using PRO-seq to determine how evolutionary changes at enhancers regulate target genes in mammals
- Our paper describing our Unified Probabilistic Modeling Framework
- Our paper describing a probabilistic modeling framework to examine transcription initiation and promoter-proximal pausing in human cells
Pandemic-related work
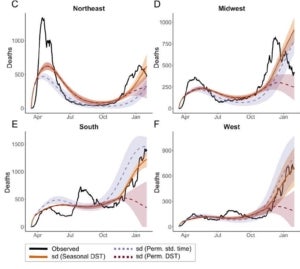
In a recent project led by Ziyi Mo in collaboration with Rob Martienssen’s lab, we explored whether circadian immunity contributes significantly to seasonality of respiratory viruses, including influenza and SARS-CoV-2. Following the general Susceptibility-Infection-Recovery-Susceptibility (SIRS) paradigm, we developed models for both influenza and COVID-19, and fitted them to public data for infections (in influenza) and hospitalizations and deaths (in COVID-19). Interestingly, these models suggest that local sunrise time is a better predictor of the basic reproductive number (R0) than climate, even when day length is taken into account. Moreover, the models predict a window of susceptibility when local sunrise time corresponds to the morning commute and contact rate is expected to be high. We predict that retaining daylight savings time in the fall would reduce the length of this window, and substantially reduce seasonal waves of respiratory infections.
Inspired by the possibility that SARS-CoV-2 may have been transmitted to humans from bats, Dick McCombie’s lab recently sequenced the genomes of the Jamaican fruit bat (Artibeus jamaicensis) and the Mesoamerican mustached bat (Pteronotus mesoamericanus) using the Oxford Nanopore Technologies long-read platform. They were also interested in studying the unusual longevity and cancer resistance of bats. Working closely with the McCombie lab, postdoc Armin Scheben led a broad comparative analysis of these two new bat genomes together with 13 additional bat genomes and the genomes of several other mammals. Armin identified a number of unusual properties of immune-related genes that may shed light on their exceptional tolerance of viral infection and/or longevity. For example, he found that the critical type I interferon locus was contracted by eight genes in the most recent common ancestor of bats, and that many antiviral genes stimulated by type I interferons were rapidly evolving. He also found evidence of positive selection on several tumor suppressors and DNA-repair genes. Altogether, this study provided valuable new genomic resources and suggested a number of potential areas for further study of the extraordinary adaptations of bats.
Funding:
Most of the research described on this page has been supported by NIH grants GM127070 and HG010346, NSF grants 1555769 and 1555754, the Cold Spring Harbor Laboratory Cancer Center, and the Simons Center for Quantitative Biology.